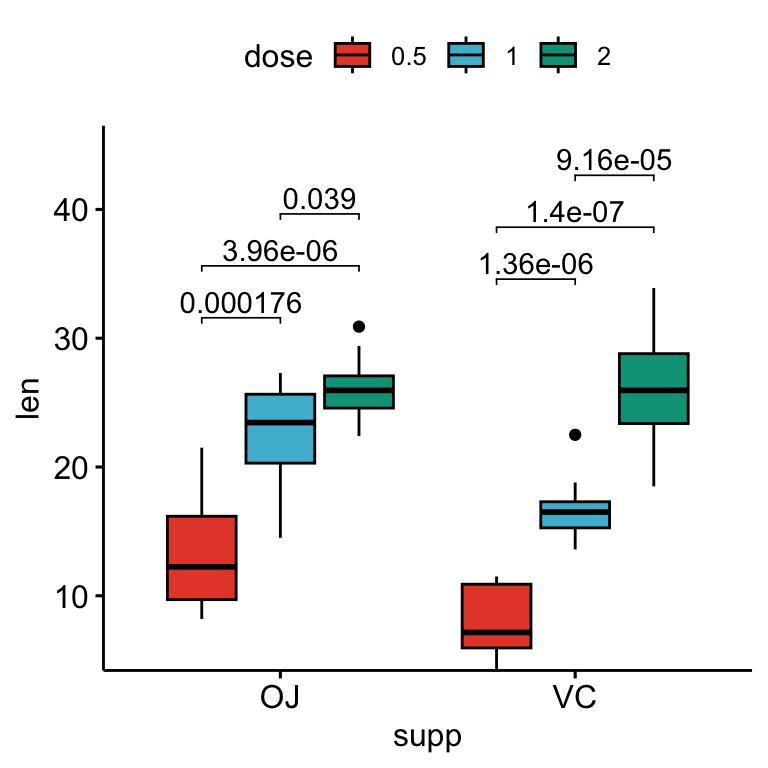
How to Add PValues onto a Grouped GGPLOT using the GGPUBR R Package - Here is the data set: P1 + geom_text( data = pval_df, aes(label = label), hjust = 1, vjust = 1,. How to install the ggpval library. When i want to add p values into my plots: It tells us the proportion of variance explained by the model.an r² of 0.80 implies that 80% of the variability in the dependent variable. You should also read this: Foreign Private Issuer Test
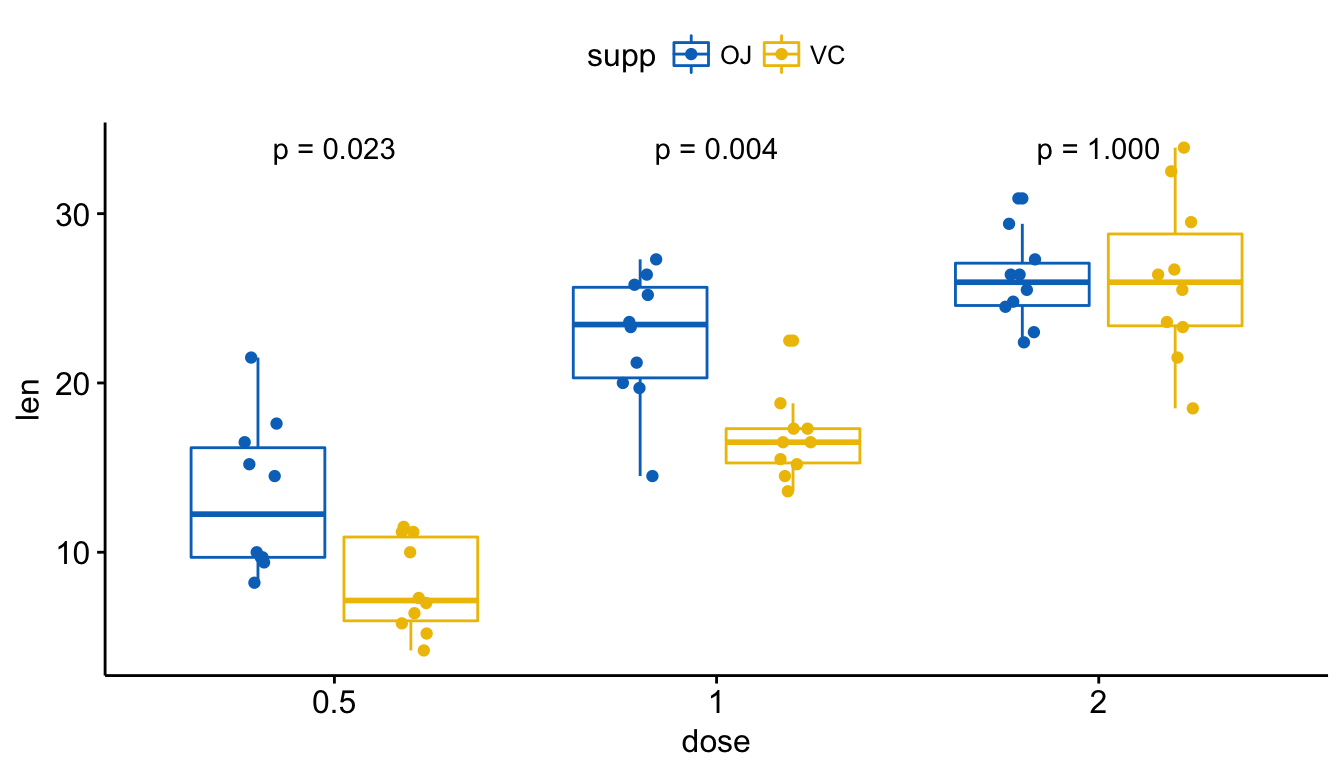
Add Pvalues and Significance Levels to ggplots Rbloggers - Dput (cauloq_datmannot) structure (list (v1 = structure (c (1l, 1l, 1l, 2l, 2l, 2l, 3l, 3l,. In this article, we’ll describe how to easily i) compare means of two or multiple groups; P + annotate(text, x = 0.5, y = 23, label = ns) + annotate(text, x = 1.5, y = 30, label = **) + annotate(text, x = 2.5,. You should also read this: Can You Have A Positive Pregnancy Test After Miscarriage

How to add the pvalue to the ggplot charts Predictive Hacks - When i want to add p values into my plots: P + annotate(text, x = 0.5, y = 23, label = ns) + annotate(text, x = 1.5, y = 30, label = **) + annotate(text, x = 2.5, y =. Dput (cauloq_datmannot) structure (list (v1 = structure (c (1l, 1l, 1l, 2l, 2l, 2l, 3l, 3l,. P1 + geom_text( data. You should also read this: Pioneer In I.q. Testing

r ggplot2 how to add lines and pvalues on a grouped barplot - An easy way could be to add manually the notations: In this article, we’ll describe how to easily i) compare means of two or multiple groups; P1 + geom_text( data = pval_df, aes(label = label), hjust = 1, vjust = 1,. Dput (cauloq_datmannot) structure (list (v1 = structure (c (1l, 1l, 1l, 2l, 2l, 2l, 3l, 3l,. Rstatix::group_by(modules) %>% rstatix::t_test(values. You should also read this: Ishihara 14 Plate Test
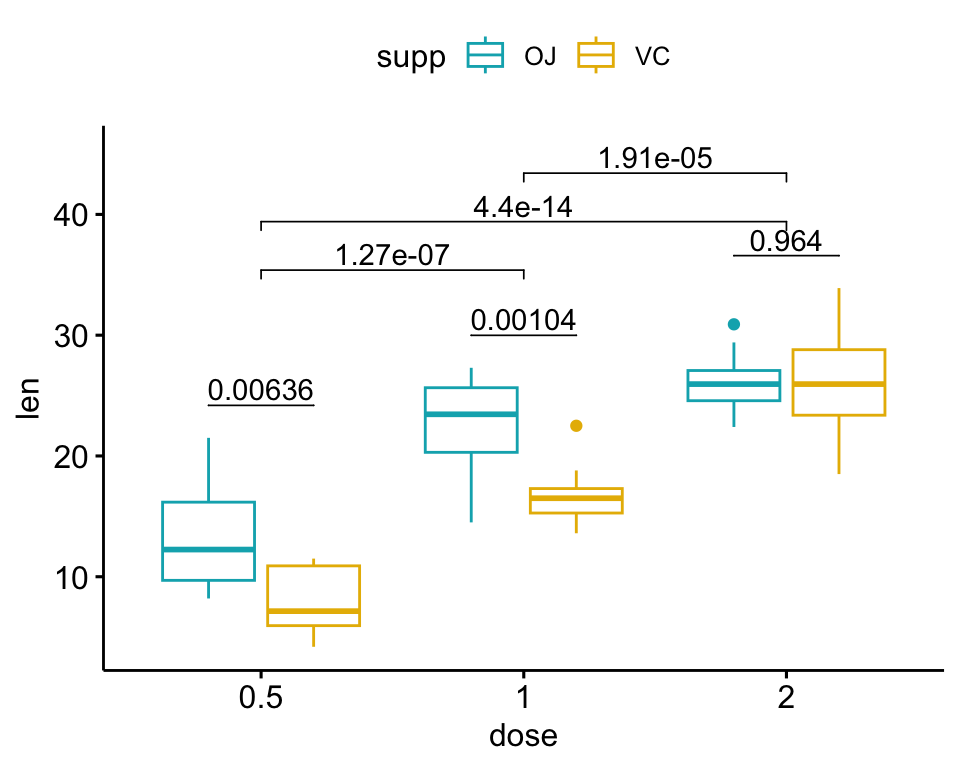
How to Add PValues onto a Grouped GGPLOT using the GGPUBR R Package - You need to specify x = 'phenotypes' in add_xy_position rather than x = 'values': Frequently asked questions are available on datanovia ggpubr faq page, for example: Dput (cauloq_datmannot) structure (list (v1 = structure (c (1l, 1l, 1l, 2l, 2l, 2l, 3l, 3l,. Examples, containing two and three groups by x position, are shown. P1 + geom_text( data = pval_df, aes(label. You should also read this: Does Blue Lotus Pop On A Drug Test
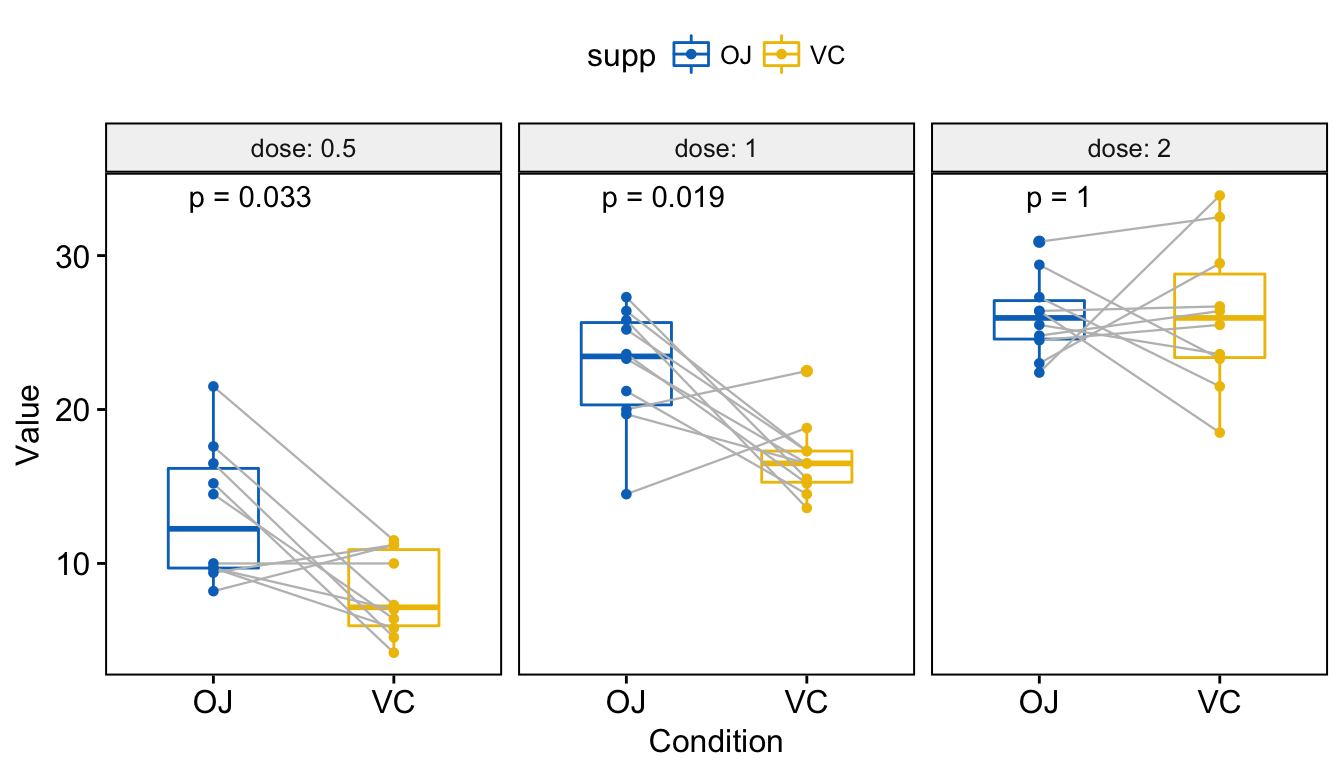
Add Pvalues and Significance Levels to ggplots Rbloggers - We will follow the steps below for adding significance levels onto a ggplot: P1 + geom_text( data = pval_df, aes(label = label), hjust = 1, vjust = 1,. It tells us the proportion of variance explained by the model.an r² of 0.80 implies that 80% of the variability in the dependent variable is explained by the model. How to install. You should also read this: Western Blot Herpes Test
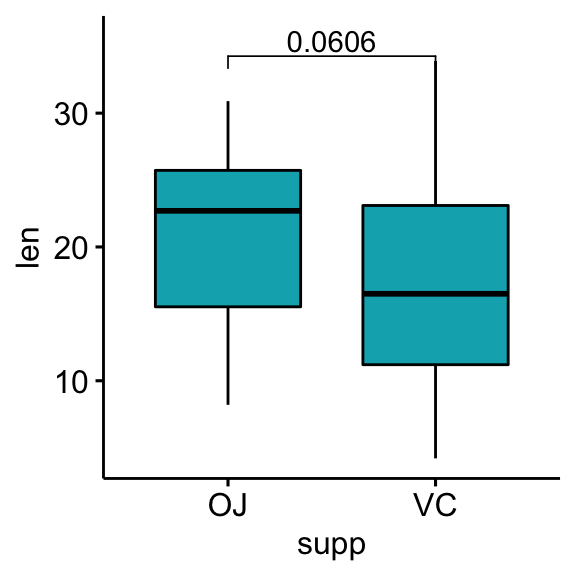
How to Add PValues onto Basic GGPLOTS Datanovia - When i want to add p values into my plots: In this article, we’ll describe how to easily i) compare means of two or multiple groups; P1 + geom_text( data = pval_df, aes(label = label), hjust = 1, vjust = 1,. An easy way could be to add manually the notations: It tells us the proportion of variance explained by. You should also read this: Drug Residue Test Kit

r Add p value for ggplot2 with ANOVA three or more groups data - Dput (cauloq_datmannot) structure (list (v1 = structure (c (1l, 1l, 1l, 2l, 2l, 2l, 3l, 3l,. P1 + geom_text( data = pval_df, aes(label = label), hjust = 1, vjust = 1,. When i want to add p values into my plots: Examples, containing two and three groups by x position, are shown. Frequently asked questions are available on datanovia ggpubr. You should also read this: Fda Animal Testing
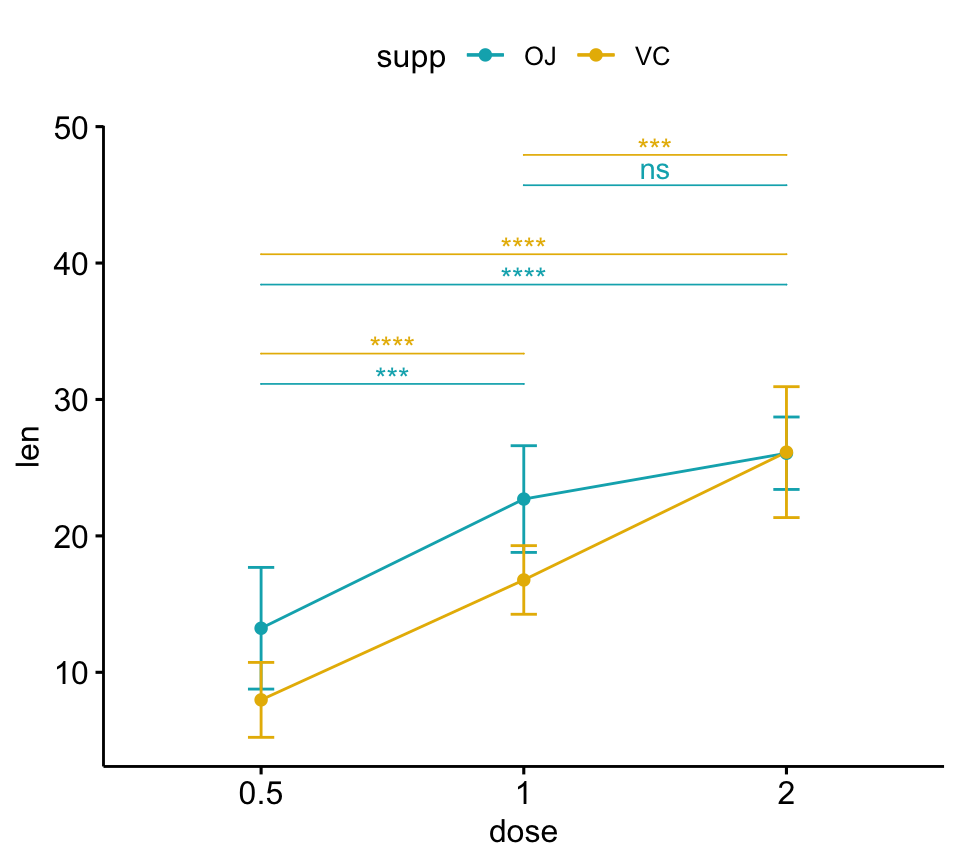
How to Add PValues onto a Grouped GGPLOT using the GGPUBR R Package - Frequently asked questions are available on datanovia ggpubr faq page, for example: P + annotate(text, x = 0.5, y = 23, label = ns) + annotate(text, x = 1.5, y = 30, label = **) + annotate(text, x = 2.5, y =. We will follow the steps below for adding significance levels onto a ggplot: In this article, we’ll describe. You should also read this: Baka To Test To Shoukanjuu Hideyoshi
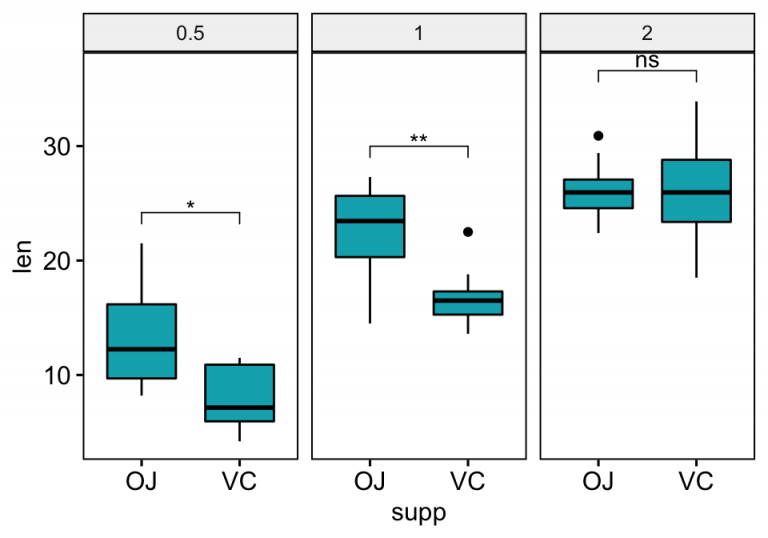
How to Add Pvalues to GGPLOT Facets Step by Step Guide Datanovia - How to install the ggpval library. We will follow the steps below for adding significance levels onto a ggplot: It tells us the proportion of variance explained by the model.an r² of 0.80 implies that 80% of the variability in the dependent variable is explained by the model. An easy way could be to add manually the notations: Rstatix::group_by(modules) %>%. You should also read this: Apo E Genotype Testing